28 Load gene names from a text file
Ryabhatta
It is possible to load gene names from a text file with a .csv or .txt format. To load genes, select the Load gene names from file tab, then use the Load genes button. This will provide an option to load the gene file using Browse.. button. Once file loading bar shows “Upload complete”, select Load the file button.
The gene names are case sensitive. The gene name format has to match the species to which the data belongs. For example, Homo sapiens gene names need to be all capital letters, while Mus musculus gene names are mostly proper case.
It is also important to note that some common protein names are not the same as the gene name. For example, CD134 is encoded by the gene TNFRSF4 in humans.
Finally, gene homologs in different species do not have the same characters. The mouse homolog of human OCT4 is Pou5f1. Just changing the format from upper case to proper case will not work to identify the homolog in this case.
Loading gene names from a file
If gene names were entered manually, they can be saved for use in posterity using the Save genes under the Load gene names from file tab. The saved genes can be recalled to be examined in another Seurat object or later during the same analysis.
Gene names saved using the Save genes can be recalled only within the current session. If the app is closed, the saved genes are lost.
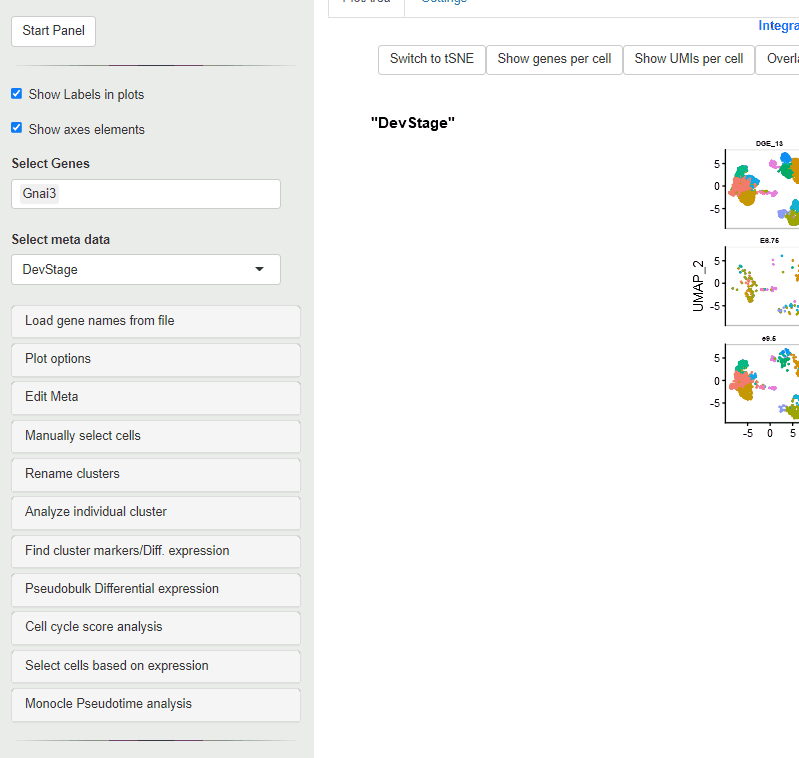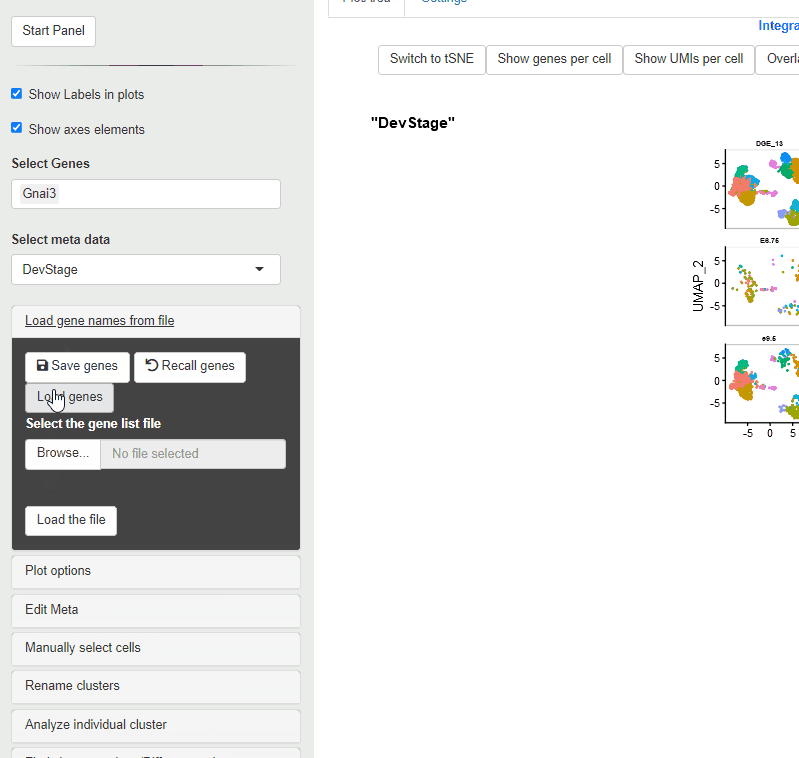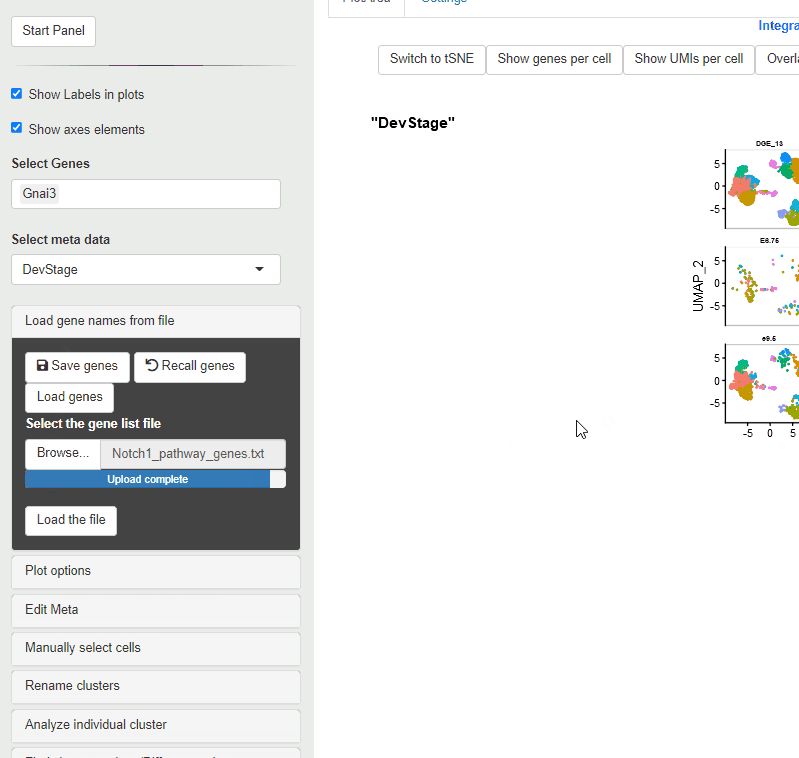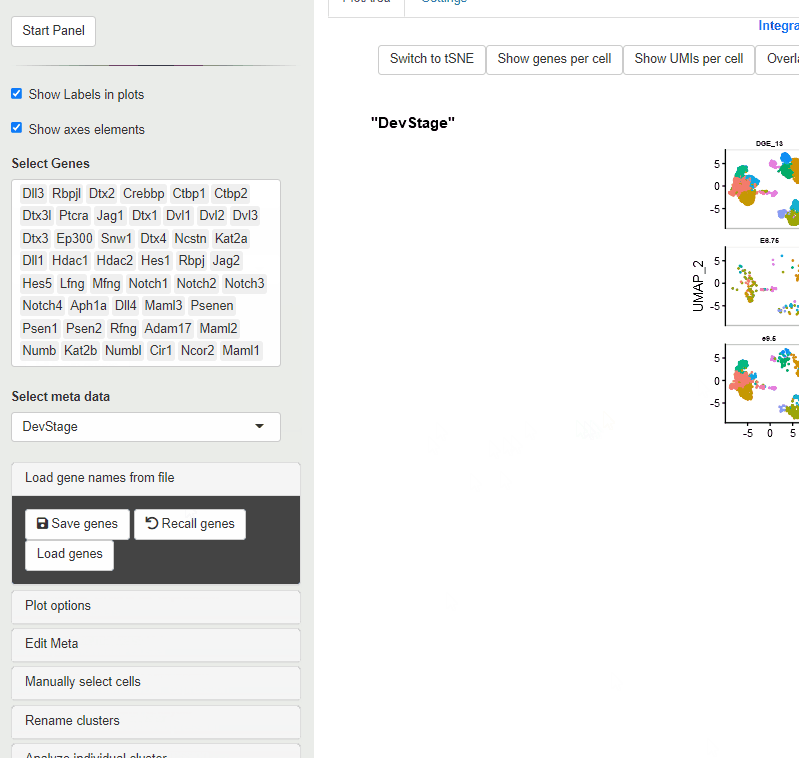
Using this approach, upto 50 genes can be loaded at one time
During the loading process, gene names that are present in the Seurat file are retained.