32 Analyze Individual Clusters
Ryabhatta
Analyzing an individual cluster or condition from a larger Seurat object is possible in Ryabhatta. To perform sub-cluster or sub-category analysis
- Ensure the cells/cluster that is of interest is set as identity.
- This can be achieved by Select the meta data category and assign it as Identities (see Assign meta information as identity) OR use the IdentVersion tool to select a previously used identities OR use select cells (manually or using expression threshold) and rename the cells as new cluster
- Select Analyze Individual Cluster panel
- Select the identities you wish to subset (You can choose multiple identities)
- Choose the number of PCs (dims) to perform the initial analysis
- Select Preview
When you select preview, the Seurat object is subset to the selected identities. Then a new seurat object is created that only contains the cells from the selected identities and genes that are at least expressed in 3 or more of these selected cells.
Next, normalization, data scaling and principal component analysis and a UMAP dimensionality reduction are done on this Seurat object.
Note If the object is a integrated Seurat object, the integrated assay is used to perform this sub-cluster analysis.
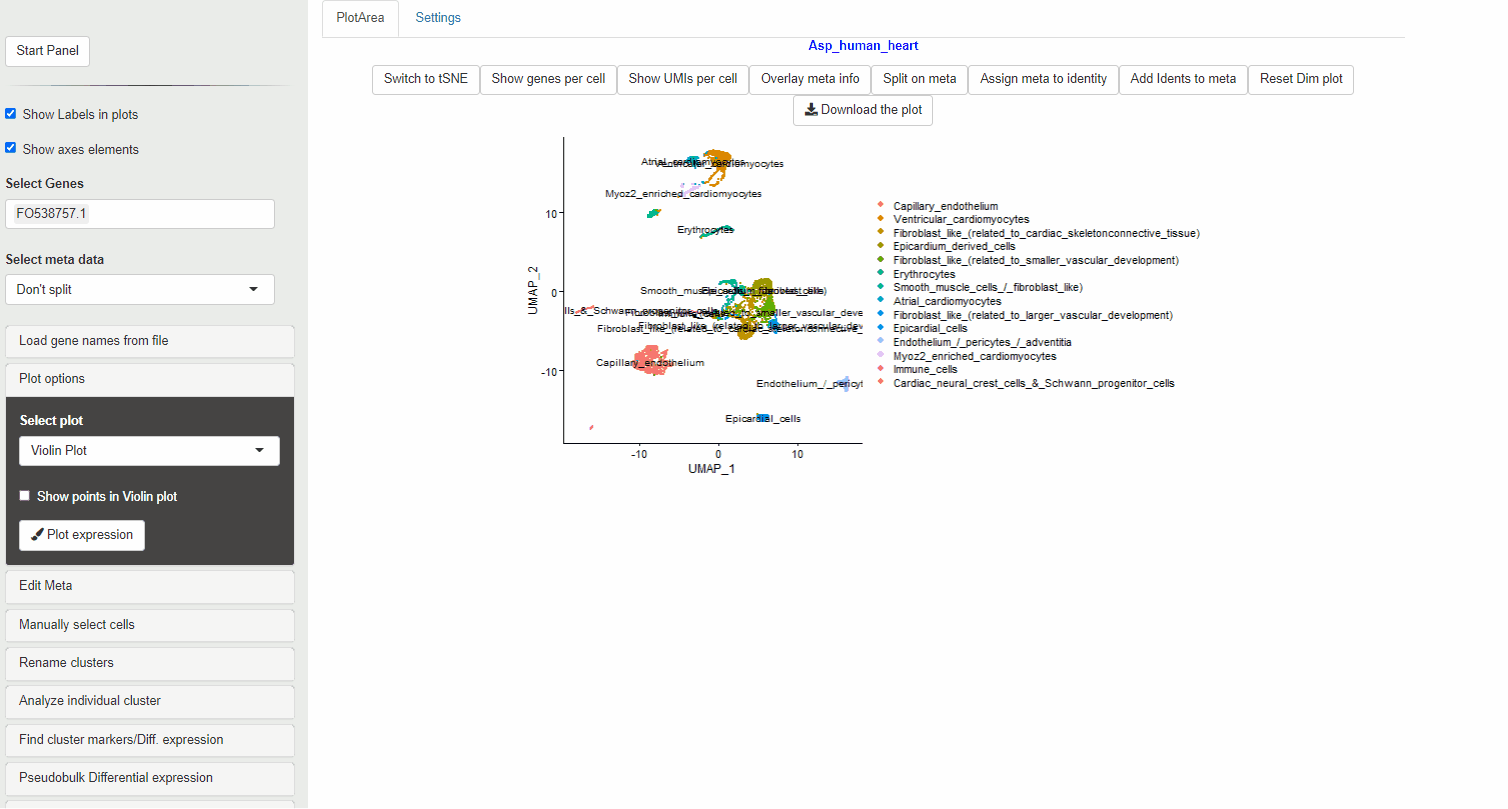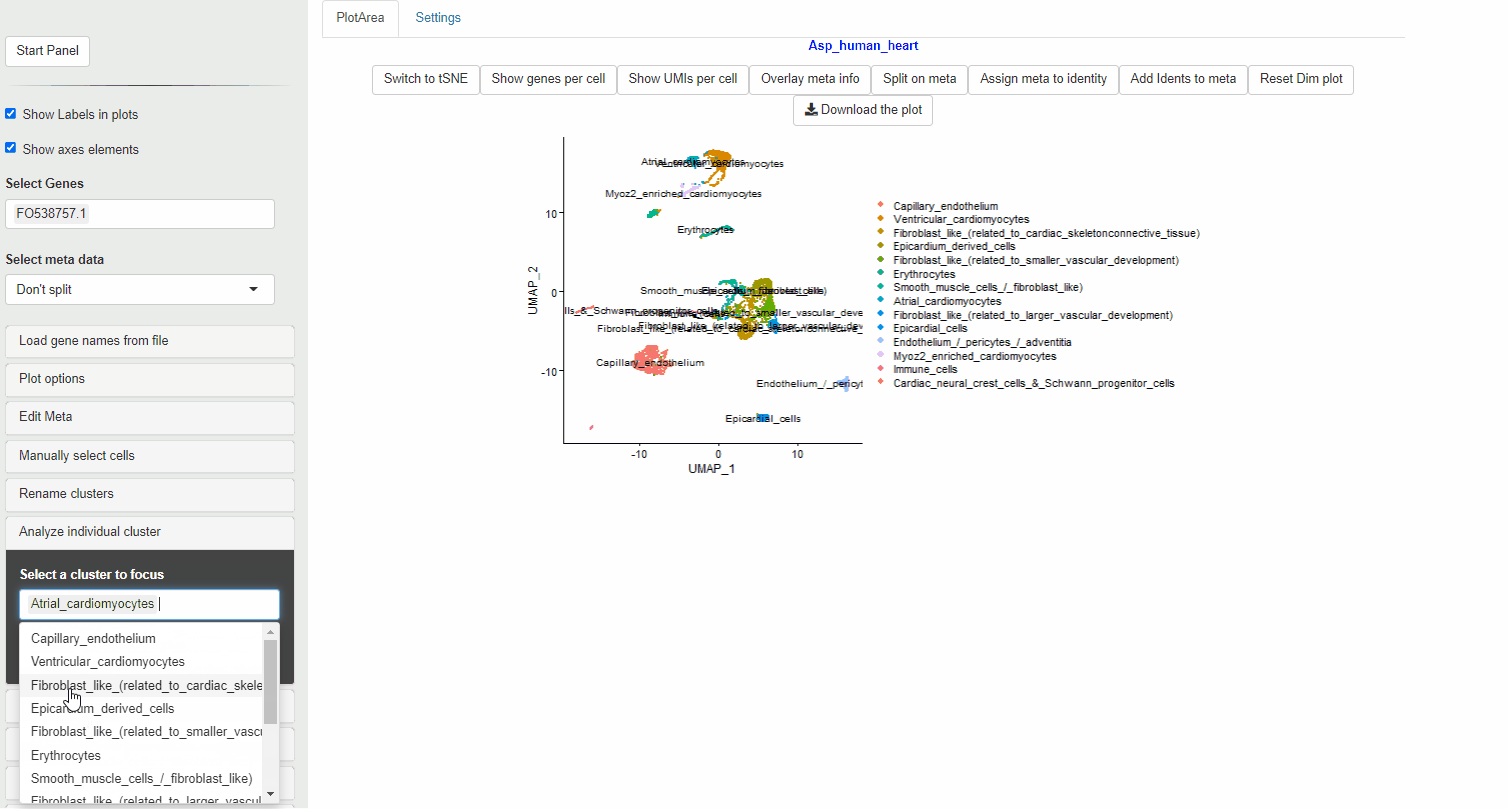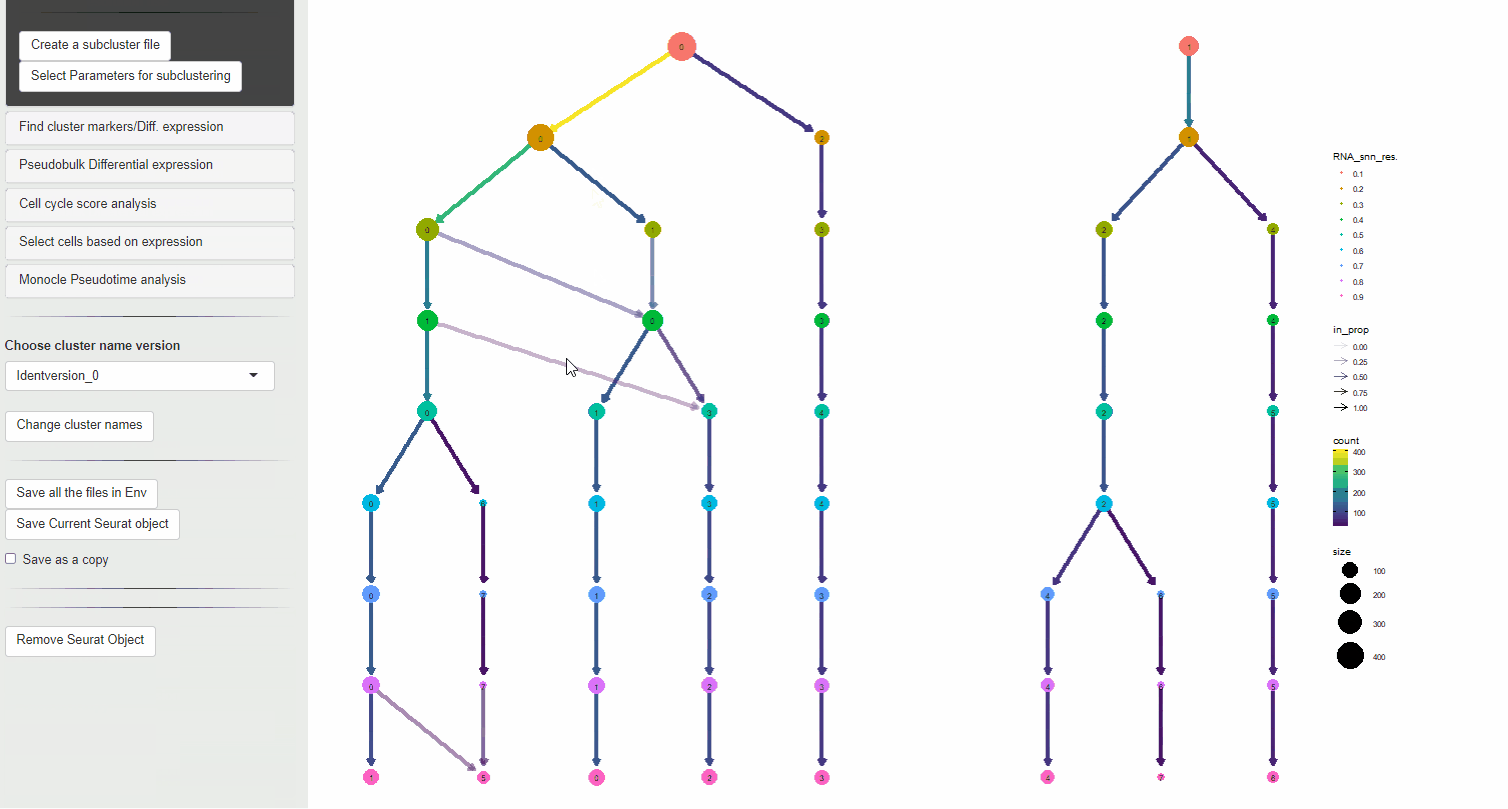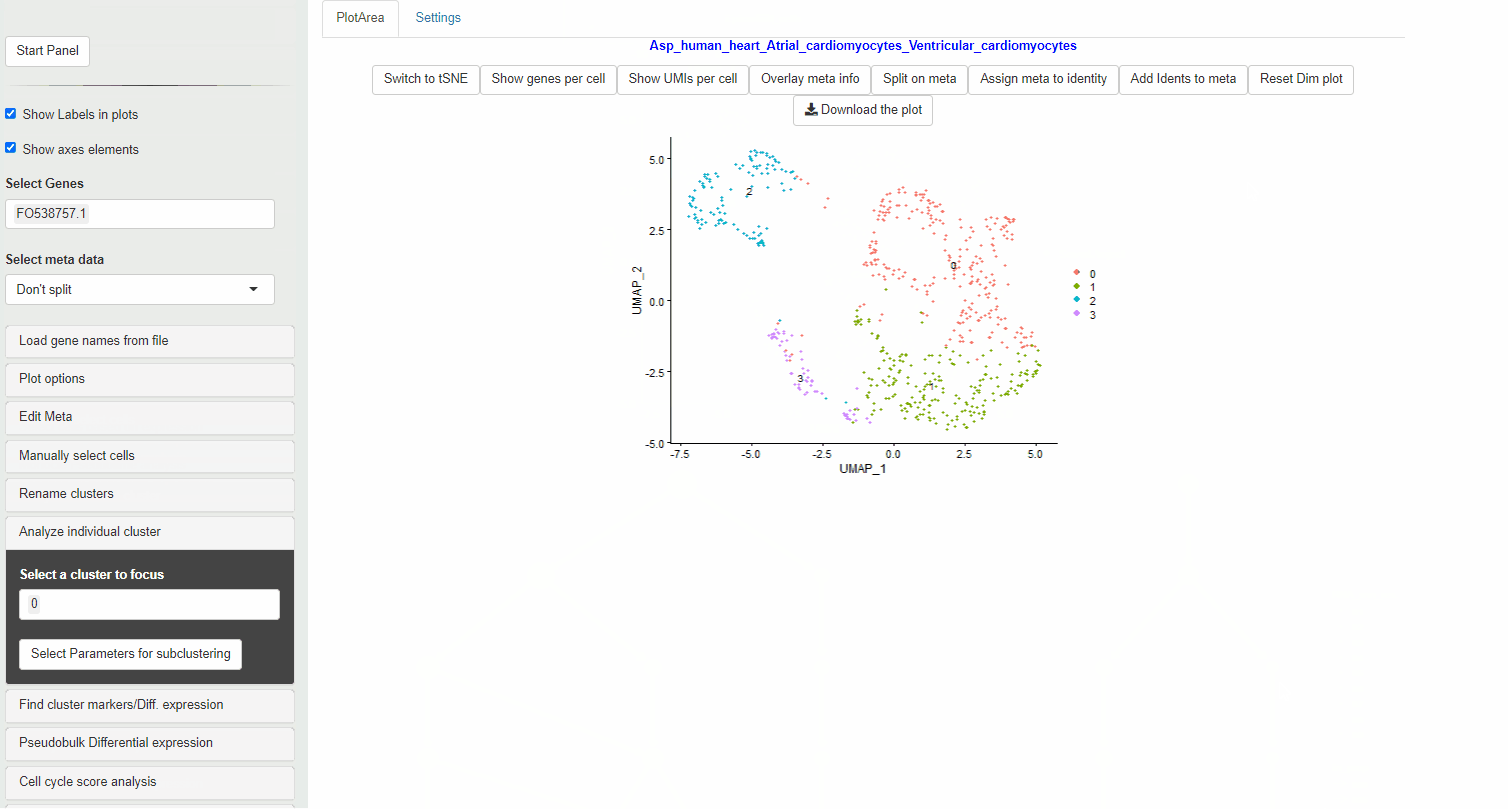
Then, a clustering is performed using multiple resolution values using the Louvain clustering method, resulting in multiple sets of clusters with varying levels of separation of clusters (broad –> granular). A clusTree diagram is then drawn using these clusters showing the size of individual cluster (number of cells in each cluster) and the number of clusters obtained at each resolution tested. This can be used to select the resolution parameter desired.
Once the dimensionality reduction plot and a clustree diagram are visible.
- Select a resolution that closely reflects the number of expected clusters
- Click Create subcluster file button
This will create a new subclutser for the selected clusters.
Analyze Individual Cluster